The base class for all user-defined models. More...
#include <BCModel.h>
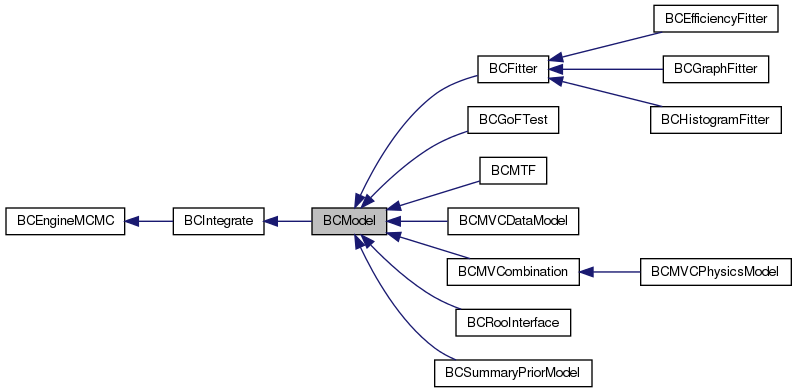
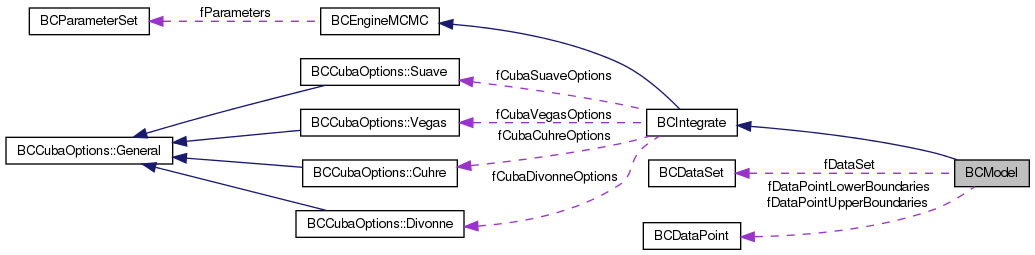
Public Member Functions | |
Constructors and destructors | |
| BCModel (const char *name="model") | |
| BCModel (const BCModel &bcmodel) | |
| virtual | ~BCModel () |
Assignment operators | |
| BCModel & | operator= (const BCModel &bcmodel) |
Member functions (get) | |
| const std::string & | GetName () const |
| double | GetModelAPrioriProbability () const |
| double | GetModelAPosterioriProbability () const |
| BCDataSet * | GetDataSet () const |
| BCDataPoint * | GetDataPointLowerBoundaries () const |
| BCDataPoint * | GetDataPointUpperBoundaries () const |
| double | GetDataPointLowerBoundary (unsigned int index) const |
| double | GetDataPointUpperBoundary (unsigned int index) const |
| bool | GetFlagBoundaries () const |
| unsigned | GetNDataPoints () const |
| BCDataPoint * | GetDataPoint (unsigned int index) const |
Member functions (set) | |
| void | SetName (const char *name) |
| void | SetModelAPrioriProbability (double probability) |
| void | SetModelAPosterioriProbability (double probability) |
| virtual int | AddParameter (BCParameter *parameter) |
| void | SetDataSet (BCDataSet *dataset) |
| void | SetSingleDataPoint (BCDataPoint *datapoint) |
| void | SetSingleDataPoint (BCDataSet *dataset, unsigned int index) |
| void | SetDataBoundaries (unsigned int index, double lowerboundary, double upperboundary, bool fixed=false) |
| void | SetDataPointLowerBoundaries (BCDataPoint *datasetlowerboundaries) |
| void | SetDataPointUpperBoundaries (BCDataPoint *datasetupperboundaries) |
| void | SetDataPointLowerBoundary (int index, double lowerboundary) |
| void | SetDataPointUpperBoundary (int index, double upperboundary) |
| int | SetPrior (int index, TF1 *f) |
| int | SetPrior (const char *name, TF1 *f) |
| int | SetPriorDelta (int index, double value) |
| int | SetPriorDelta (const char *name, double value) |
| int | SetPriorGauss (int index, double mean, double sigma) |
| int | SetPriorGauss (const char *name, double mean, double sigma) |
| int | SetPriorGauss (int index, double mean, double sigmadown, double sigmaup) |
| int | SetPriorGauss (const char *name, double mean, double sigmadown, double sigmaup) |
| int | SetPrior (int index, TH1 *h, bool flag=false) |
| int | SetPrior (const char *name, TH1 *h, bool flag=false) |
| int | SetPriorConstant (int index) |
| int | SetPriorConstant (const char *name) |
| int | SetPriorConstantAll () |
Member functions (miscellaneous methods) | |
| void | Copy (const BCModel &bcmodel) |
| double | APrioriProbability (const std::vector< double > ¶meters) |
| virtual double | LogAPrioriProbability (const std::vector< double > ¶meters) |
| virtual double | Likelihood (const std::vector< double > ¶ms) |
| virtual double | LogLikelihood (const std::vector< double > ¶ms)=0 |
| double | ProbabilityNN (const std::vector< double > ¶ms) |
| double | LogProbabilityNN (const std::vector< double > ¶meters) |
| double | Probability (const std::vector< double > ¶meter) |
| double | LogProbability (const std::vector< double > ¶meter) |
| virtual double | SamplingFunction (const std::vector< double > ¶meters) |
| double | Eval (const std::vector< double > ¶meters) |
| virtual double | LogEval (const std::vector< double > ¶meters) |
| virtual void | CorrelateDataPointValues (std::vector< double > &x) |
| double | GetPvalueFromChi2 (const std::vector< double > &par, int sigma_index) |
| double | GetPvalueFromKolmogorov (const std::vector< double > &par, int index) |
| double | GetPvalueFromChi2NDoF (std::vector< double > par, int sigma_index) |
| BCH1D * | CalculatePValue (std::vector< double > par, bool flag_histogram=false) |
| double | GetPValue () |
| double | GetPValueNDoF () |
| double | GetChi2NDoF () |
| std::vector< double > | GetChi2Runs (int dataIndex, int sigmaIndex) |
| void | SetGoFNIterationsMax (int n) |
| void | SetGoFNIterationsRun (int n) |
| void | SetGoFNChains (int n) |
| double | HessianMatrixElement (const BCParameter *parameter1, const BCParameter *parameter2, std::vector< double > point) |
| void | PrintSummary () |
| void | PrintResults (const char *file) |
| void | PrintShortFitSummary (int chi2flag=0) |
| void | PrintHessianMatrix (std::vector< double > parameters) |
| virtual double | CDF (const std::vector< double > &, int, bool) |
 Public Member Functions inherited from BCIntegrate Public Member Functions inherited from BCIntegrate | |
| BCIntegrate () | |
| BCIntegrate (const BCIntegrate &bcintegrate) | |
| virtual | ~BCIntegrate () |
| BCIntegrate & | operator= (const BCIntegrate &bcintegrate) |
| int | ReadMarginalizedFromFile (const char *file) |
| BCH1D * | GetMarginalized (const BCParameter *parameter) |
| BCH1D * | GetMarginalized (const char *name) |
| BCH1D * | GetMarginalized (unsigned index) |
| BCH2D * | GetMarginalized (const BCParameter *parameter1, const BCParameter *parameter2) |
| BCH2D * | GetMarginalized (const char *name1, const char *name2) |
| BCH2D * | GetMarginalized (unsigned index1, unsigned index2) |
| int | PrintAllMarginalized1D (const char *filebase) |
| int | PrintAllMarginalized2D (const char *filebase) |
| int | PrintAllMarginalized (const char *file, std::string options1d="BTsiB3CS1D0pdf0Lmeanmode", std::string options2d="BTfB3CS1meangmode", unsigned int hdiv=1, unsigned int ndiv=1) |
| double | GetIntegral () const |
| BCIntegrate::BCOptimizationMethod | GetOptimizationMethod () const |
| BCIntegrate::BCIntegrationMethod | GetIntegrationMethod () const |
| BCIntegrate::BCMarginalizationMethod | GetMarginalizationMethod () const |
| BCIntegrate::BCSASchedule | GetSASchedule () const |
| void | GetRandomVectorUnitHypercube (std::vector< double > &x) const |
| void | GetRandomVectorInParameterSpace (std::vector< double > &x) const |
| double | GetRandomPoint (std::vector< double > &x) |
| int | GetNIterationsMin () const |
| int | GetNIterationsMax () const |
| int | GetNIterationsPrecisionCheck () const |
| int | GetNIterationsOutput () const |
| int | GetNIterations () const |
| double | GetRelativePrecision () const |
| double | GetAbsolutePrecision () const |
| BCCubaMethod | GetCubaIntegrationMethod () const |
| const BCCubaOptions::Vegas & | GetCubaVegasOptions () const |
| const BCCubaOptions::Suave & | GetCubaSuaveOptions () const |
| const BCCubaOptions::Divonne & | GetCubaDivonneOptions () const |
| const BCCubaOptions::Cuhre & | GetCubaCuhreOptions () const |
| BCH1D * | GetSlice (const BCParameter *parameter, const std::vector< double > parameters=std::vector< double >(0), int bins=0, bool flag_norm=true) |
| BCH1D * | GetSlice (const char *name, const std::vector< double > parameters=std::vector< double >(0), int nbins=0, bool flag_norm=true) |
| BCH2D * | GetSlice (const BCParameter *parameter1, const BCParameter *parameter2, const std::vector< double > parameters=std::vector< double >(0), int bins=0, bool flag_norm=true) |
| BCH2D * | GetSlice (const char *name1, const char *name2, const std::vector< double > parameters=std::vector< double >(0), int nbins=0, bool flag_norm=true) |
| BCH2D * | GetSlice (unsigned index1, unsigned index2, const std::vector< double > parameters=std::vector< double >(0), int nbins=0, bool flag_norm=true) |
| double | GetError () const |
| TMinuit * | GetMinuit () |
| int | GetMinuitErrorFlag () const |
| double | GetSAT0 () const |
| double | GetSATmin () const |
| double | GetBestFitParameter (unsigned index) const |
| double | GetBestFitParameterError (unsigned index) const |
| double | GetLogMaximum () |
| const std::vector< double > & | GetBestFitParameters () const |
| const std::vector< double > & | GetBestFitParameterErrors () const |
| void | SetMinuitArlist (double *arglist) |
| void | SetFlagIgnorePrevOptimization (bool flag) |
| void | SetOptimizationMethod (BCIntegrate::BCOptimizationMethod method) |
| void | SetIntegrationMethod (BCIntegrate::BCIntegrationMethod method) |
| void | SetMarginalizationMethod (BCIntegrate::BCMarginalizationMethod method) |
| void | SetSASchedule (BCIntegrate::BCSASchedule schedule) |
| void | SetNIterationsMin (int niterations) |
| void | SetNIterationsMax (int niterations) |
| void | SetNIterationsPrecisionCheck (int niterations) |
| void | SetNIterationsOutput (int niterations) |
| void | SetRelativePrecision (double relprecision) |
| void | SetAbsolutePrecision (double absprecision) |
| void | SetCubaIntegrationMethod (BCCubaMethod type) |
| void | SetCubaOptions (const BCCubaOptions::Vegas &options) |
| void | SetCubaOptions (const BCCubaOptions::Suave &options) |
| void | SetCubaOptions (const BCCubaOptions::Divonne &options) |
| void | SetCubaOptions (const BCCubaOptions::Cuhre &options) |
| void | SetSAT0 (double T0) |
| void | SetSATmin (double Tmin) |
| void | SetFlagWriteSAToFile (bool flag) |
| TTree * | GetSATree () |
| void | InitializeSATree () |
| double | Normalize () |
| double | Integrate (BCIntegrationMethod intmethod) |
| double | Integrate () |
| double | Integrate (BCIntegrationMethod type, tRandomizer randomizer, tEvaluator evaluator, tIntegralUpdater updater, std::vector< double > &sums) |
| double | EvaluatorMC (std::vector< double > &sums, const std::vector< double > &point, bool &accepted) |
| int | MarginalizeAll () |
| int | MarginalizeAll (BCMarginalizationMethod margmethod) |
| virtual void | MarginalizePreprocess () |
| virtual void | MarginalizePostprocess () |
| void | SAInitialize () |
| std::vector< double > | FindMode (std::vector< double > start=std::vector< double >()) |
| std::vector< double > | FindMode (BCIntegrate::BCOptimizationMethod optmethod, std::vector< double > start=std::vector< double >()) |
| double | SATemperature (double t) |
| double | SATemperatureBoltzmann (double t) |
| double | SATemperatureCauchy (double t) |
| virtual double | SATemperatureCustom (double t) |
| std::vector< double > | GetProposalPointSA (const std::vector< double > &x, int t) |
| std::vector< double > | GetProposalPointSABoltzmann (const std::vector< double > &x, int t) |
| std::vector< double > | GetProposalPointSACauchy (const std::vector< double > &x, int t) |
| virtual std::vector< double > | GetProposalPointSACustom (const std::vector< double > &x, int t) |
| std::vector< double > | SAHelperGetRandomPointOnHypersphere () |
| double | SAHelperGetRadialCauchy () |
| double | SAHelperSinusToNIntegral (int dim, double theta) |
| virtual void | ResetResults () |
| std::string | DumpIntegrationMethod (BCIntegrationMethod type) |
| std::string | DumpCurrentIntegrationMethod () |
| std::string | DumpUsedIntegrationMethod () |
| std::string | DumpMarginalizationMethod (BCMarginalizationMethod type) |
| std::string | DumpCurrentMarginalizationMethod () |
| std::string | DumpUsedMarginalizationMethod () |
| std::string | DumpOptimizationMethod (BCOptimizationMethod type) |
| std::string | DumpCurrentOptimizationMethod () |
| std::string | DumpUsedOptimizationMethod () |
| std::string | DumpCubaIntegrationMethod (BCCubaMethod type) |
| std::string | DumpCubaIntegrationMethod () |
| void | SetBestFitParameters (const std::vector< double > &x) |
| void | SetBestFitParameters (const std::vector< double > &x, const double &new_value, double &old_value) |
| unsigned | GetNIntegrationVariables () |
| double | CalculateIntegrationVolume () |
| bool | CheckMarginalizationAvailability (BCMarginalizationMethod type) |
| bool | CheckMarginalizationIndices (TH1 *hist, const std::vector< unsigned > &index) |
 Public Member Functions inherited from BCEngineMCMC Public Member Functions inherited from BCEngineMCMC | |
| void | WriteMarkovChain (bool flag) |
| BCEngineMCMC () | |
| BCEngineMCMC (const BCEngineMCMC &enginemcmc) | |
| virtual | ~BCEngineMCMC () |
| BCEngineMCMC & | operator= (const BCEngineMCMC &engineMCMC) |
| unsigned | MCMCGetNChains () const |
| unsigned | MCMCGetNLag () const |
| const std::vector< unsigned > & | MCMCGetNIterations () const |
| unsigned | MCMCGetCurrentIteration () const |
| unsigned | MCMCGetCurrentChain () const |
| unsigned | MCMCGetNIterationsConvergenceGlobal () const |
| bool | MCMCGetFlagConvergenceGlobal () const |
| unsigned | MCMCGetNIterationsMax () const |
| unsigned | MCMCGetNIterationsRun () const |
| unsigned | MCMCGetNIterationsPreRunMin () const |
| unsigned | MCMCGetNIterationsUpdate () const |
| unsigned | MCMCGetNIterationsUpdateMax () const |
| std::vector< int > | MCMCGetNTrialsTrue () const |
| int | MCMCGetNTrials () const |
| const std::vector< double > & | MCMCGetprobMean () const |
| const std::vector< double > & | MCMCGetVariance () const |
| const std::vector< double > & | MCMCGetTrialFunctionScaleFactor () const |
| std::vector< double > | MCMCGetTrialFunctionScaleFactor (unsigned ichain) const |
| double | MCMCGetTrialFunctionScaleFactor (unsigned ichain, unsigned ipar) |
| const std::vector< double > & | MCMCGetx () const |
| std::vector< double > | MCMCGetx (unsigned ichain) |
| double | MCMCGetx (unsigned ichain, unsigned ipar) const |
| const std::vector< double > & | MCMCGetLogProbx () const |
| double | MCMCGetLogProbx (unsigned ichain) |
| int | MCMCGetPhase () const |
| const std::vector< double > & | MCMCGetMaximumPoints () const |
| std::vector< double > | MCMCGetMaximumPoint (unsigned i) const |
| const std::vector< double > & | MCMCGetMaximumLogProb () const |
| int | MCMCGetFlagInitialPosition () const |
| double | MCMCGetRValueCriterion () const |
| double | MCMCGetRValueParametersCriterion () const |
| double | MCMCGetRValue () const |
| double | MCMCGetRValueParameters (unsigned i) |
| bool | MCMCGetRValueStrict () const |
| bool | MCMCGetFlagRun () const |
| TTree * | MCMCGetMarkovChainTree (unsigned i) |
| BCH1D * | MCMCGetH1Marginalized (unsigned i) |
| BCH2D * | MCMCGetH2Marginalized (unsigned i, unsigned j) |
| BCParameter * | GetParameter (int index) const |
| BCParameter * | GetParameter (const char *name) const |
| unsigned int | GetNParameters () const |
| unsigned int | GetNFixedParameters () |
| unsigned int | GetNFreeParameters () |
| const std::vector< double > & | GetBestFitParametersMarginalized () const |
| void | MCMCSetTrialFunctionScaleFactor (std::vector< double > scale) |
| void | MCMCSetNChains (unsigned n) |
| void | MCMCSetNLag (unsigned n) |
| void | MCMCSetNIterationsMax (unsigned n) |
| void | MCMCSetNIterationsRun (unsigned n) |
| void | MCMCSetNIterationsPreRunMin (unsigned n) |
| void | MCMCSetNIterationsUpdate (unsigned n) |
| void | MCMCSetNIterationsUpdateMax (unsigned n) |
| void | MCMCSetMinimumEfficiency (double efficiency) |
| void | MCMCSetMaximumEfficiency (double efficiency) |
| void | MCMCSetRandomSeed (unsigned seed) |
| void | MCMCSetWriteChainToFile (bool flag) |
| void | MCMCSetWritePreRunToFile (bool flag) |
| void | MCMCSetInitialPositions (const std::vector< double > &x0s) |
| void | MCMCSetInitialPositions (std::vector< std::vector< double > > x0s) |
| void | MCMCSetFlagInitialPosition (int flag) |
| void | MCMCSetFlagOrderParameters (bool flag) |
| void | MCMCSetFlagFillHistograms (bool flag) |
| void | MCMCSetFlagPreRun (bool flag) |
| void | MCMCSetRValueCriterion (double r) |
| void | MCMCSetRValueParametersCriterion (double r) |
| void | MCMCSetRValueStrict (bool strict=true) |
| void | MCMCSetMarkovChainTrees (const std::vector< TTree * > &trees) |
| void | MCMCInitializeMarkovChainTrees () |
| int | SetMarginalized (unsigned index, TH1D *h) |
| int | SetMarginalized (unsigned index1, unsigned index2, TH2D *h) |
| void | MCMCSetValuesDefault () |
| void | MCMCSetValuesQuick () |
| void | MCMCSetValuesDetail () |
| void | MCMCSetPrecision (BCEngineMCMC::Precision precision) |
| void | SetNbins (unsigned int nbins) |
| void | Copy (const BCEngineMCMC &enginemcmc) |
| virtual int | AddParameter (const char *name, double min, double max, const char *latexname="") |
| virtual void | MCMCTrialFunction (unsigned ichain, std::vector< double > &x) |
| virtual double | MCMCTrialFunctionSingle (unsigned ichain, unsigned ipar) |
| bool | MCMCGetProposalPointMetropolis (unsigned chain, std::vector< double > &x) |
| bool | MCMCGetProposalPointMetropolis (unsigned chain, unsigned parameter, std::vector< double > &x) |
| bool | MCMCGetNewPointMetropolis (unsigned chain=0) |
| bool | MCMCGetNewPointMetropolis (unsigned chain, unsigned parameter) |
| void | MCMCInChainCheckMaximum () |
| void | MCMCInChainUpdateStatistics () |
| void | MCMCInChainFillHistograms () |
| void | MCMCInChainTestConvergenceAllChains () |
| void | MCMCInChainWriteChains () |
| int | MCMCMetropolis () |
| int | MCMCMetropolisPreRun () |
| void | MCMCResetRunStatistics () |
| void | MCMCInitializeMarkovChains () |
| int | MCMCInitialize () |
| void | ClearParameters (bool hard=false) |
| virtual void | MCMCIterationInterface () |
| virtual void | MCMCUserIterationInterface () |
| virtual void | MCMCCurrentPointInterface (std::vector< double > &, int, bool) |
Private Member Functions | |
| int | CompareStrings (const char *string1, const char *string2) |
| BCDataPoint * | VectorToDataPoint (const std::vector< double > &data) |
Additional Inherited Members | |
 Public Types inherited from BCIntegrate Public Types inherited from BCIntegrate | |
| enum | BCOptimizationMethod { kOptEmpty, kOptSimAnn, kOptMetropolis, kOptMinuit, kOptDefault, NOptMethods } |
| enum | BCIntegrationMethod { kIntEmpty, kIntMonteCarlo, kIntCuba, kIntGrid, kIntDefault, NIntMethods } |
| enum | BCMarginalizationMethod { kMargEmpty, kMargMetropolis, kMargMonteCarlo, kMargGrid, kMargDefault, NMargMethods } |
| enum | BCSASchedule { kSACauchy, kSABoltzmann, kSACustom, NSAMethods } |
| enum | BCCubaMethod { kCubaVegas, kCubaSuave, kCubaDivonne, kCubaCuhre, NCubaMethods } |
| typedef void(BCIntegrate::* | tRandomizer )(std::vector< double > &) const |
| typedef double(BCIntegrate::* | tEvaluator )(std::vector< double > &, const std::vector< double > &, bool &) |
| typedef void(* | tIntegralUpdater )(const std::vector< double > &, const int &, double &, double &) |
 Static Public Member Functions inherited from BCIntegrate Static Public Member Functions inherited from BCIntegrate | |
| static void | IntegralUpdaterMC (const std::vector< double > &sums, const int &nIterations, double &integral, double &absprecision) |
| static int | CubaIntegrand (const int *ndim, const double xx[], const int *ncomp, double ff[], void *userdata) |
| static void | FCNLikelihood (int &npar, double *grad, double &fval, double *par, int flag) |
 Protected Member Functions inherited from BCIntegrate Protected Member Functions inherited from BCIntegrate | |
| unsigned | IntegrationOutputFrequency () const |
| void | LogOutputAtStartOfIntegration (BCIntegrationMethod type, BCCubaMethod cubatype) |
| void | LogOutputAtIntegrationStatusUpdate (BCIntegrationMethod type, double integral, double absprecision, int nIterations) |
| void | LogOutputAtEndOfIntegration (double integral, double absprecision, double relprecision, int nIterations) |
| void | Copy (const BCIntegrate &bcintegrate) |
Detailed Description
The base class for all user-defined models.
- Version
- 1.0
- Date
- 08.2008 This class represents a model. It contains a container of prior distributions and the likelihood. The methods that implement the prior and the likelihood have to be overloaded by the user in the user defined model class derived from this class.
Constructor & Destructor Documentation
| BCModel::BCModel | ( | const char * | name = "model") |
| BCModel::BCModel | ( | const BCModel & | bcmodel) |
The copy constructor.
Definition at line 56 of file BCModel.cxx.
|
virtual |
Member Function Documentation
|
virtual |
Adds a parameter to the model.
- Parameters
-
parameter A model parameter
- See Also
- AddParameter(const char * name, double lowerlimit, double upperlimit);
Reimplemented from BCEngineMCMC.
Definition at line 225 of file BCModel.cxx.
| double BCModel::APrioriProbability | ( | const std::vector< double > & | parameters) |
Returns the prior probability.
- Parameters
-
parameters A set of parameter values
- Returns
- The prior probability p(parameters)
- See Also
- GetPrior(std::vector<double> parameters)
Definition at line 267 of file BCModel.cxx.
| BCH1D * BCModel::CalculatePValue | ( | std::vector< double > | par, |
| bool | flag_histogram = false |
||
| ) |
Definition at line 451 of file BCModel.cxx.
|
inlinevirtual |
1dim cumulative distribution function of the probability to get the data f(x_i|param) for a single measurement, assumed to be of identical functional form for all measurements
- Parameters
-
parameters The parameter values at which point to compute the cdf index The data point index starting at 0,1...N-1 lower only needed for discrete distributions! Return the CDF for the count one less than actually observed, e.g. in Poisson process, if 3 actually observed, then CDF(2) is returned
Reimplemented in BCHistogramFitter, and BCGraphFitter.
|
private |
Compares to strings
Definition at line 1094 of file BCModel.cxx.
| void BCModel::Copy | ( | const BCModel & | bcmodel) |
|
virtual |
|
virtual |
Overloaded function to evaluate integral.
Reimplemented from BCIntegrate.
Definition at line 330 of file BCModel.cxx.
|
inline |
| std::vector< double > BCModel::GetChi2Runs | ( | int | dataIndex, |
| int | sigmaIndex | ||
| ) |
For a Gaussian problem, calculate the chi2 of the longest run of consecutive values above/below the expected values
- Parameters
-
dataIndex component of datapoint with the observed value sigmaIndex component of datapoint with uncertainty
Definition at line 368 of file BCModel.cxx.
| BCDataPoint * BCModel::GetDataPoint | ( | unsigned int | index) | const |
- Parameters
-
index The index of the data point.
- Returns
- The data point in the current data set at index
Definition at line 132 of file BCModel.cxx.
|
inline |
| double BCModel::GetDataPointLowerBoundary | ( | unsigned int | index) | const |
- Parameters
-
index The index of the variable.
- Returns
- The lower boundary of possible data values for a particular variable.
Definition at line 142 of file BCModel.cxx.
|
inline |
| double BCModel::GetDataPointUpperBoundary | ( | unsigned int | index) | const |
- Parameters
-
index The index of the variable.
- Returns
- The upper boundary of possible data values for a particular variable.
Definition at line 148 of file BCModel.cxx.
|
inline |
| bool BCModel::GetFlagBoundaries | ( | ) | const |
Checks if the boundaries have been defined
- Returns
- true, if the boundaries have been set, false otherwise
Definition at line 154 of file BCModel.cxx.
|
inline |
|
inline |
|
inline |
| unsigned BCModel::GetNDataPoints | ( | ) | const |
- Returns
- The number of data points in the current data set.
Definition at line 123 of file BCModel.cxx.
|
inline |
| double BCModel::GetPvalueFromChi2 | ( | const std::vector< double > & | par, |
| int | sigma_index | ||
| ) |
Calculate p-value from Chi2 distribution for Gaussian problems
- Parameters
-
par Parameter set for the calculation of the likelihood sigma_index Index of the sigma/uncertainty for the data points (for data in format "x y erry" the index would be 2)
Definition at line 351 of file BCModel.cxx.
| double BCModel::GetPvalueFromChi2NDoF | ( | std::vector< double > | par, |
| int | sigma_index | ||
| ) |
Definition at line 375 of file BCModel.cxx.
| double BCModel::GetPvalueFromKolmogorov | ( | const std::vector< double > & | par, |
| int | index | ||
| ) |
Calculate p-value from Kolmogorov-Smirnov test statistic for 1D - datasets.
- Parameters
-
par Parameter set for the calculation of the likelihood index Index of the data point in the BCDataSet (for data in format "x y erry" the index would be 1)
Definition at line 394 of file BCModel.cxx.
| double BCModel::HessianMatrixElement | ( | const BCParameter * | parameter1, |
| const BCParameter * | parameter2, | ||
| std::vector< double > | point | ||
| ) |
Calculates the matrix element of the Hessian matrix
- Parameters
-
parameter1 The parameter for the first derivative parameter2 The parameter for the first derivative
- Returns
- The matrix element of the Hessian matrix
Definition at line 501 of file BCModel.cxx.
|
virtual |
Returns the likelihood
- Parameters
-
params A set of parameter values
- Returns
- The likelihood
Definition at line 324 of file BCModel.cxx.
|
virtual |
Returns natural logarithm of the prior probability. Method needs to be overloaded by the user.
- Parameters
-
parameters A set of parameter values
- Returns
- The prior probability p(parameters)
- See Also
- GetPrior(std::vector<double> parameters)
Reimplemented in BCGoFTest, BCMVCDataModel, BCSummaryPriorModel, and BCRooInterface.
Definition at line 273 of file BCModel.cxx.
|
virtual |
Overloaded function to evaluate integral.
Reimplemented from BCIntegrate.
Definition at line 336 of file BCModel.cxx.
|
pure virtual |
Calculates natural logarithm of the likelihood. Method needs to be overloaded by the user.
- Parameters
-
params A set of parameter values
- Returns
- Natural logarithm of the likelihood
Implemented in BCMTF, BCEfficiencyFitter, BCMVCombination, BCHistogramFitter, BCGraphFitter, BCGoFTest, BCMVCDataModel, BCSummaryPriorModel, BCMVCPhysicsModel, and BCRooInterface.
| double BCModel::LogProbability | ( | const std::vector< double > & | parameter) |
Returns natural logarithm of the a posteriori probability given a set of parameter values
- Parameters
-
parameters A set of parameter values
- Returns
- The a posteriori probability
Definition at line 255 of file BCModel.cxx.
|
inline |
Defaut assignment operator
Definition at line 115 of file BCModel.cxx.
| void BCModel::PrintHessianMatrix | ( | std::vector< double > | parameters) |
Prints matrix elements of the Hessian matrix
- Parameters
-
parameters The parameter values at which point to evaluate the matrix
Definition at line 1057 of file BCModel.cxx.
| void BCModel::PrintResults | ( | const char * | file) |
Prints a summary of the Markov Chain Monte Carlo to a file.
Definition at line 816 of file BCModel.cxx.
| void BCModel::PrintShortFitSummary | ( | int | chi2flag = 0) |
Prints a short summary of the fit results on the screen.
Definition at line 1030 of file BCModel.cxx.
| void BCModel::PrintSummary | ( | ) |
Prints a summary on the screen.
Definition at line 772 of file BCModel.cxx.
| double BCModel::Probability | ( | const std::vector< double > & | parameter) |
Returns the a posteriori probability given a set of parameter values
- Parameters
-
parameters A set of parameter values
- Returns
- The a posteriori probability
Definition at line 249 of file BCModel.cxx.
| double BCModel::ProbabilityNN | ( | const std::vector< double > & | params) |
Returns the likelihood times prior probability given a set of parameter values
- Parameters
-
params A set of parameter values
- Returns
- The likelihood times prior probability
Definition at line 243 of file BCModel.cxx.
|
virtual |
Sampling function used for importance sampling. Method needs to be overloaded by the user.
- Parameters
-
parameters A set of parameter values
- Returns
- The probability density at the parameter values
Definition at line 342 of file BCModel.cxx.
| void BCModel::SetDataBoundaries | ( | unsigned int | index, |
| double | lowerboundary, | ||
| double | upperboundary, | ||
| bool | fixed = false |
||
| ) |
Definition at line 194 of file BCModel.cxx.
|
inline |
| void BCModel::SetDataPointLowerBoundary | ( | int | index, |
| double | lowerboundary | ||
| ) |
Sets the lower boundary of possible data values for a particular variable
Definition at line 546 of file BCModel.cxx.
|
inline |
| void BCModel::SetDataPointUpperBoundary | ( | int | index, |
| double | upperboundary | ||
| ) |
Sets the upper boundary of possible data values for a particular variable
Definition at line 552 of file BCModel.cxx.
|
inline |
|
inline |
|
inline |
|
inline |
|
inline |
|
inline |
|
inline |
| int BCModel::SetPrior | ( | int | index, |
| TF1 * | f | ||
| ) |
Set prior for a parameter.
- Parameters
-
index The parameter index f A pointer to a function describing the prior
- Returns
- An error code.
Definition at line 558 of file BCModel.cxx.
| int BCModel::SetPrior | ( | const char * | name, |
| TF1 * | f | ||
| ) |
Set prior for a parameter.
- Parameters
-
name The parameter name f A pointer to a function describing the prior
- Returns
- An error code.
Definition at line 579 of file BCModel.cxx.
| int BCModel::SetPrior | ( | int | index, |
| TH1 * | h, | ||
| bool | flag = false |
||
| ) |
Set prior for a parameter.
- Parameters
-
index parameter index h pointer to a histogram describing the prior flag whether or not to use linear interpolation
- Returns
- An error code.
Definition at line 682 of file BCModel.cxx.
| int BCModel::SetPrior | ( | const char * | name, |
| TH1 * | h, | ||
| bool | flag = false |
||
| ) |
Set prior for a parameter.
- Parameters
-
name parameter name h pointer to a histogram describing the prior flag whether or not to use linear interpolation
- Returns
- An error code.
Definition at line 719 of file BCModel.cxx.
| int BCModel::SetPriorConstant | ( | int | index) |
Set constant prior for this parameter
- Parameters
-
index the index of the parameter
- Returns
- An error code
Definition at line 732 of file BCModel.cxx.
|
inline |
| int BCModel::SetPriorConstantAll | ( | ) |
Enable caching the constant value of the prior, so LogAPrioriProbability is called only once. Note that the prior for ALL parameters is assumed to be constant. The value is computed from the parameter ranges, so make sure these are defined before this method is called.
- Returns
- An error code
Definition at line 753 of file BCModel.cxx.
| int BCModel::SetPriorDelta | ( | int | index, |
| double | value | ||
| ) |
Set delta-function prior for a parameter. Note: this sets the parameter range to the specified value. The old parameter range is lost.
- Parameters
-
index The parameter index value The position of the delta function.
- Returns
- An error code.
Definition at line 592 of file BCModel.cxx.
| int BCModel::SetPriorDelta | ( | const char * | name, |
| double | value | ||
| ) |
Set delta-function prior for a parameter. Note: this sets the parameter range to the specified value. The old parameter range is lost.
- Parameters
-
name The parameter name value The position of the delta function.
- Returns
- An error code.
Definition at line 602 of file BCModel.cxx.
| int BCModel::SetPriorGauss | ( | int | index, |
| double | mean, | ||
| double | sigma | ||
| ) |
Set Gaussian prior for a parameter.
- Parameters
-
index The parameter index mean The mean of the Gaussian sigma The sigma of the Gaussian
- Returns
- An error code.
Definition at line 610 of file BCModel.cxx.
| int BCModel::SetPriorGauss | ( | const char * | name, |
| double | mean, | ||
| double | sigma | ||
| ) |
Set Gaussian prior for a parameter.
- Parameters
-
name The parameter name mean The mean of the Gaussian sigma The sigma of the Gaussian
- Returns
- An error code.
Definition at line 631 of file BCModel.cxx.
| int BCModel::SetPriorGauss | ( | int | index, |
| double | mean, | ||
| double | sigmadown, | ||
| double | sigmaup | ||
| ) |
Set Gaussian prior for a parameter with two different widths.
- Parameters
-
index The parameter index mean The mean of the Gaussian sigmadown The sigma (down) of the Gaussian sigmaup The sigma (up)of the Gaussian
- Returns
- An error code.
Definition at line 644 of file BCModel.cxx.
| int BCModel::SetPriorGauss | ( | const char * | name, |
| double | mean, | ||
| double | sigmadown, | ||
| double | sigmaup | ||
| ) |
Set Gaussian prior for a parameter with two different widths.
- Parameters
-
name The parameter name mean The mean of the Gaussian sigmadown The sigma (down) of the Gaussian sigmaup The sigma (up)of the Gaussian
- Returns
- An error code.
Definition at line 669 of file BCModel.cxx.
| void BCModel::SetSingleDataPoint | ( | BCDataPoint * | datapoint) |
Sets a single data point as data set.
- Parameters
-
datapoint A data point
Definition at line 172 of file BCModel.cxx.
| void BCModel::SetSingleDataPoint | ( | BCDataSet * | dataset, |
| unsigned int | index | ||
| ) |
Definition at line 185 of file BCModel.cxx.
|
private |
Member Data Documentation
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
The documentation for this class was generated from the following files:
 1.8.4
1.8.4